Recently, the BORG has been used to wrap the Berkeley Advanced Reconstruction Toolbox (BART) into a GPI node library for the purpose of allowing GPI users to easily explore and utilize this new compressed sensing library. This GPI-BART library is accompanied by some example networks that demonstrate basic wavelet based compressed sensing as well as some of the advanced joint compressed sensing and parallel imaging techniques that the BART package provides for MR reconstruction. This post gives an introductory overview of how the BART was wrapped in GPI and how the GPI-BART node library can be installed using the latest GPI v1.
The BORG
One of the new features in GPI
v1 is the BORG interface which allows node developers to easily assimilate
command-line tools for native use in GPI. The BORG, which stands for Building
Outside Relationships with GPI, provides a simple wrapper interface that takes
care of the file I/O required to communicate with external binaries. In the
case of the BART, most of the tools require an input data file to process and
subsequently produce a data file as an output. The BORG simply manages the
temporary files required for this I/O. Reader and writer functions are
required to translate the Python object (in GPI) to the appropriate file format
for the command-line tool. In this case the BART comes with a Python library
that translates their CFL/HDR file format to Numpy complex float arrays. This
can be observed in the following code snippet of a GPI node that wraps the
BART's traj command.
# load commandline tools
from bart.gpi.borg import IFilePath, OFilePath, Command
from bart.python.cfl import cfl # BART file format
...
# grab user input from UI widgets
x = self.getVal('readout samples')
y = self.getVal('phase encoding lines')
a = self.getVal('acceleration')
r = self.getVal('radial')
g = self.getVal('golden-ratio sampling')
# assemble the argument string
args = [base_path+'/traj']
args += ['-x '+str(x)]
args += ['-y '+str(y)]
args += ['-a '+str(a)]
args += r*['-r']
args += g*['-G']
args += d*['-D']
# setup temp file for getting data back
# from the external command
out = OFilePath(cfl.readcfl, asuffix=['.cfl','.hdr'])
args += [out]
# run commandline and echo full command string
print(Command(*args))
# set GPI node output
self.setData('trajectory', out.data())
return 0
The example above starts with a local import of the BORG tools, which were
developed in tandem with this node library. The GPI widgets values are then
translated to command-line arguments, which are held as strings in a Python
list object. Since the Traj node generates k-space trajectory coordinates,
it requires an output file path to write out the trajectory data. The
OFilePath object takes a reference to the file format reader function and
generates a random temporary filename which is included in the arguments list.
When the Command object executes with the argument list, it spawns a traj
process and waits for it to finish and then it uses the reader function to
convert the file to a Numpy array. The OFilePath object provides a reference
to the Numpy via its data() method. Finally, when the OFilePath object
falls out of scope it cleans up the temporary file on disk. The random
filenames are important since the GPI canvas may contain multiple instances of
the Traj node. The BORG interface simplifies this process which can save
time when wrapping a large library like the BART.
For more examples, check out the wrapper nodes to the BART and FSL libraries.
Why a Fork?
The GPI-BART library is built off of a fork of the actual BART project. While the GPI wrappers could be considered a separate project, the BART project is currently undergoing a lot of development and the fork effectively ensures that the interface between the GPI wrappers and the BART is compatible. Another advantage is that this library can coexist with other BART installations on the same machine without interfering.
BART Data Conventions
The BART and GPI projects have a few key organizational differences in how they
convey numeric arrays and k-space coordinates. In order to pass the data
between nodes from each library, the data need to be manipulated to fit the
each node's requirements. This is easily accomplished with the core GPI nodes.
Lets start with an example of the worst case scenario: the traj node.
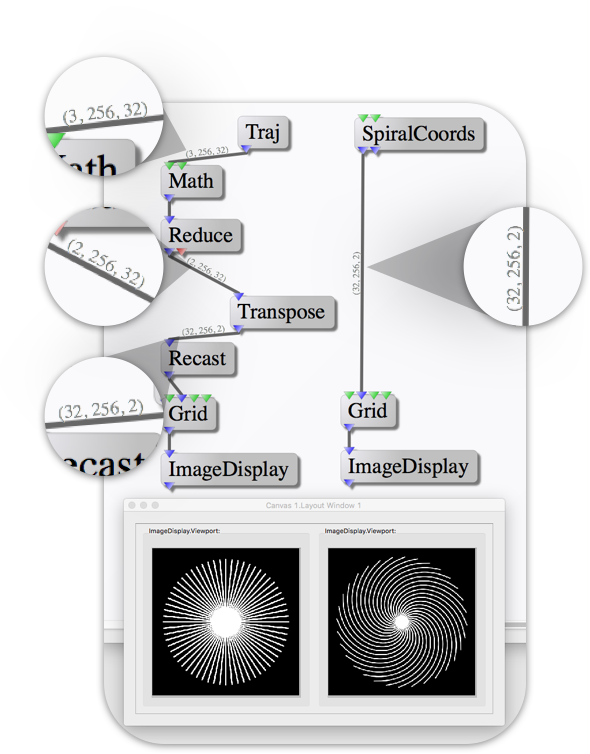
The image above shows an example gridded 2D radial trajectory (via the BART
node library) and a gridded 2D spiral trajectory (via the GPI core library).
Starting from the top nodes, you can see that the Traj node produces radial
coordinates with dimensions [3, 256, 32] which is 3 cartesian coordinates,
256 sample points and 32 radial arms. The spiral coordinates from
SpiralCoords are ordered [32, 256, 2] for 32 spiral arms, 256 sample points
and 2 cartesian coordinates.
Step 1: Reduce Coordinates
Since this is a 2D trajectory, the Traj node passes zeros for the 'Z'
coordinate and the SpiralCoords node passes 2 coordinates per point. In
order to use the radial trajectory with the core Grid node, the 3rd
coordinate of the array must be cropped out. This is accomplished with the
Reduce node set to B/E where the '(B)eginning' index is 1 and the
'(E)nding' index is 2. You can see that after Reduce, the Traj dimensions
are [2, 256, 32].
Step 2: Transpose Dimensions
The Transpose node is used to flip the order that the trajectory data is
organized in. After the Transpose node, the dimensions are [32, 256, 2],
which matches the order of the spiral coordinates.
Step 3: Recast the Data Type
If you hover the mouse cursor over the coordinate port on the Grid node a
tooltip will pop up with the data type required by that port. In this case,
Grid can take floating point arrays with single or double precision.
Hovering over the Traj output port shows that the array is of type complex
float. In this case the trajectory definition only makes use of the real
component so the imaginary component is set to zero. This can be directly
recast into a float array without loss of data.
Step 4: Coordinate Scaling
You have noticed that the first node after Traj is Math. This is because
the trajectory coordinate convention used in the BART is from N/2-1 to N/2
where N is the number of points along a dimension of the grid matrix. The GPI
core library uses the convention of -0.5 to 0.5. So the coordinates are
divided by the target matrix size via element-wise scalar division.
This example has shown how the BART and core GPI nodes can be easily adapted to communicate data between the two libraries using the stock data manipulation nodes in the core library. The conventions for data between these two libraries are different as would be expected between any two libraries that are developed independently. The goal for this example is to show that a cross library data transfer may require some extra attention.
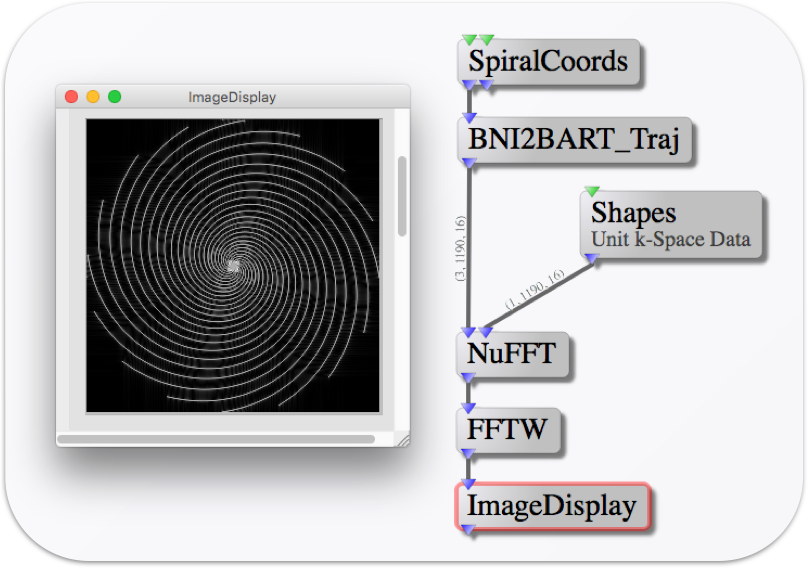
Another approach to this problem would be to integrate these differences into
the nodes themselves. An example of this can be seen in the image above. The
SpiralCoords node is generating a trajectory that is reformatted using the
BNI2BART_Traj node (included in the wrapper library). This allows the
coordinates to be directly used in the BART's NuFFT node.
GPI-BART Installation
There are two components to the GPI-BART node library installation: the BART compilation and the GPI node library installation. Before starting download or clone the GPI-BART fork:
First (if you haven't already), make a 'gpi' directory in your home directory. If you're not sure why you'd do this, checkout the post Installing Node Libraries.
$ cd ~/
$ mkdir ~/gpi
$ cd ~/gpi
Next, use git to clone the node library project into a directory called 'bart'.
$ git clone https://github.com/nckz/bart.git bart
BART Compilation
The BART compilation instructions can be found in the
README.md in the
base directory of the project source code. To summarize, the BART has some
third party library dependencies that are specific to each OS (e.g. Mac
OSX). For OSX, the Xcode and the
gcc compiler must be installed before the make command can be issued. Once
you've installed the dependencies, make the BART.
$ cd ~/gpi/bart
$ make
This will make the BART command-line executables, which will be accessible from
the ~/gpi/bart directory.
Installing Node Libraries
The BART compilation instructions above basically install the BART under a GPI
node library directory structure. GPI will look at the ~/gpi/bart directory
as if it where a Python library. In our fork, we've included the GPI wrapper
nodes under the bart/gpi
directory. This fixed directory structure allows the wrapper code to maintain
the relative paths for the BART executables. Since the GPI wrappers are pure
Python, they don't require compilation. If your GPI installation is setup with
the default paths then GPI-BART library is ready to go.
If you have path modifications in your ~/.gpirc file or the nodes are not
visible in your library menu, consult the
Configuration docs or check out the
post on Installing Node Libraries.
Events
This work has been presented at the ISMRM Workshop on Data Sampling & Image Reconstruction in Sedona Arizona this last January and will be presented at the ISMRM Annual Meeting & Exhibition in Singapore this March.
Thanks
We'd like to thanks the BART developers at UC Berkeley for their help in building these nodes.